
Soybean Phenotype Prediction
A Novel Framework for Soybean Phenotype Prediction and Salient Loci Mining via Machine Learning and Interpretability Analysis
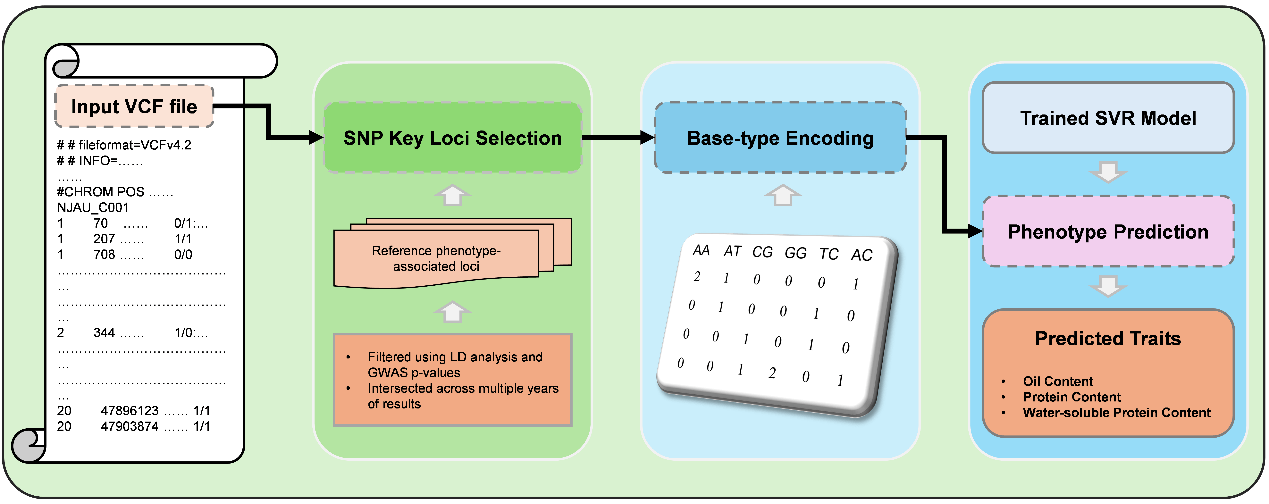
This platform uses our custom-designed Base-Type SNP encoding and a Support Vector Regression (SVR) model to predict three essential phenotypes:
- Oil Content
- Protein Content
- Water-Soluble Protein Content
Simply upload a VCF file of a soybean variety. We will extract essential SNP loci, fill in missing data using a reference genome, and deliver accurate phenotype predictions—powered by explainable machine learning.
- 🔹 Optimized for high-dimensional, small-sample genomic data
- 🔹 Auto SNP extraction and reference-based imputation
- 🔹 Accurate prediction in one click
Note:
1. Currently, only single-sample phenotype prediction is supported. The input file must be a VCF file for a single soybean variety.
2. The VCF file format can be adjusted using tools like bcftools. Please refer to the provided example file for the correct format. (Reference commands for bcftools: bcftools view -c1 -s single_sample_ID -o your_vcf_file.vcf -O v single_sample_ID.vcf)
Prediction Result:
| Oil Content Predicton | Protein Content Prediction | Water-soluble Protein Content |
|---|---|---|
| 91.88% | Unrecognized | 91.88% |